To build a multi layer perceptron model for predicting breast cancer in python.
Breast cancer. (Kaggle)
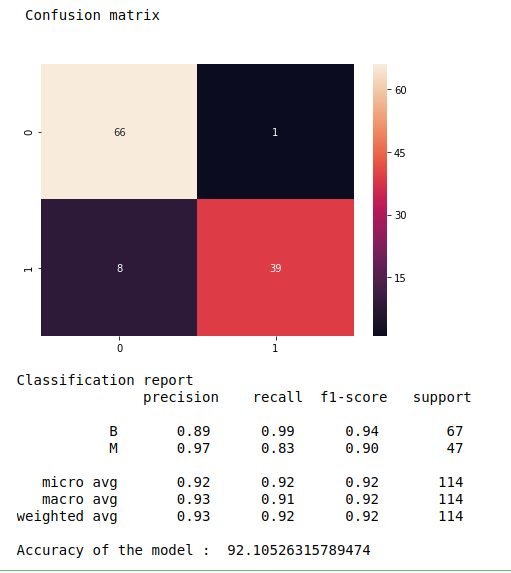
Confusion matrix, classification report and accuracy_score.
Import libraries.
Load the data set.
Fix X and y variables.
Split the data into train and test.
Build the MLP model.
Fit the train data to the model.
Predict the test data.
Evaluate the model.
Print the results.
#import necessary libraries
import warnings
warnings.filterwarnings(“ignore”)
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.neural_network import MLPClassifier
#load data
data = pd.read_csv(‘……../cancer.csv’)
#make as a data frame
df = pd.DataFrame(data)
#Checking missing values
print(“Checking missing values\n”)
print(df.isnull().sum())
#Define X and y variable
X = df.iloc[:,2:32]
y = df.iloc[:,1]
#Split train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
#Training and testing data
print(“\n”)
print(“Original data\n\n”,df.head(20))
print(“After vectorized train data\n\n”,X_train,”\n\n”,y_train)
print(“After vectorized test data\n\n”,X_test)
#Neural network model
clf = MLPClassifier(activation=’relu’,solver=’adam’, batch_size=50,alpha=1e-5,hidden_layer_sizes=(150,140,130),
random_state=42,learning_rate=’adaptive’)
#fit the model
clf.fit(X_train, y_train)
# Predicting the Test set results
y_pred = clf.predict(X_test)
# Creating the Confusion Matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
print(“\n”,”Confusion matrix\n”)
fig, ax = plt.subplots(figsize=(7,5))
sns.heatmap(cm, annot=True, fmt=’d’)
plt.show()
print(“Classification report\n”,metrics.classification_report(y_test, y_pred))
print(“Accuracy of the model : “,metrics.accuracy_score(y_test, y_pred)*100)