To classify humans based on their activity using deep neural network in R
library(keras)
Load the necessary libraries
Load the data set
Convert the categorical variables to equivalent numeric classes
Split the data set as train set and test set
Initialize the keras sequential model
Build the model with input layers,hidden layers and output layer as per the data size along with the activation function(here 1658(vectorizer count) I/P layer with relu activation and 2 O/P layer with sigmoid activation)
Compile the model with required loss,metrics and optimizer(here loss=binary_crossentropy,optimizer=adam,metrics=accuracy)
Fit the model using the train set
Predict using the test set
Evaluate the metrics
#load the required libraries
require(MASS)
library(caret)
library(keras)
#Load the data set
data=read.csv(‘/…/UCI HAR Dataset/train/X_train.txt’,header=FALSE,sep=””)
y=read.csv(‘/home/soft23/Downloads/UCI HAR Dataset/train/y_train.txt’,header=FALSE)
#To Split 80% of data as training data
smp_size train_ind #Compute the Principal component Analysis
pca_HAR summary(pca_HAR)
##Create the dataframe with PCA component and y
final_data=data.frame(x=pca_HAR$x[,1:70],y=factor(y$V1,labels=c(0:5)))
##Split the PCA dataframe for train and test
train xtrain <-train[,1:70]
ytrain <-train$y
test xtest ytest #converting the target variable to once hot encoded vectors using keras inbuilt function
train_y<-to_categorical(ytrain)
test_y #defining a keras sequential model
model %
layer_dense(units = 100, input_shape = 70 )%>%
layer_activation(activation = ‘relu’) %>%
layer_dense(units = 6)%>%
layer_activation(activation= “softmax”)
#compiling the defined model with metric = accuracy and optimiser as adam.
model %>% compile(
loss = ‘categorical_crossentropy’,
optimizer = ‘Adam’,
metrics = ‘accuracy’
)
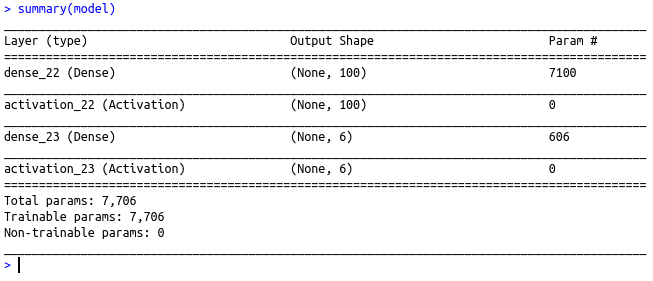
#Summary of the model
summary(model)
#fitting the model on the training dataset
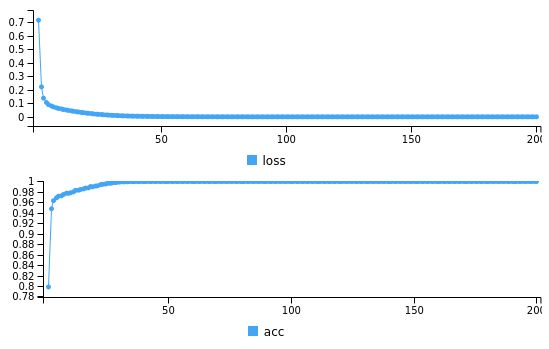
model %>%keras:: fit(as.matrix(xtrain), train_y, epochs = 200,batch_size=50)
#Predict using the Test data
yt=predict_classes(model,as.matrix(xtest))
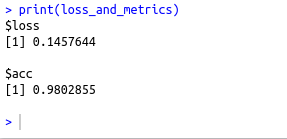
loss_and_metrics % evaluate(as.matrix(xtest), test_y)
print(loss_and_metrics)
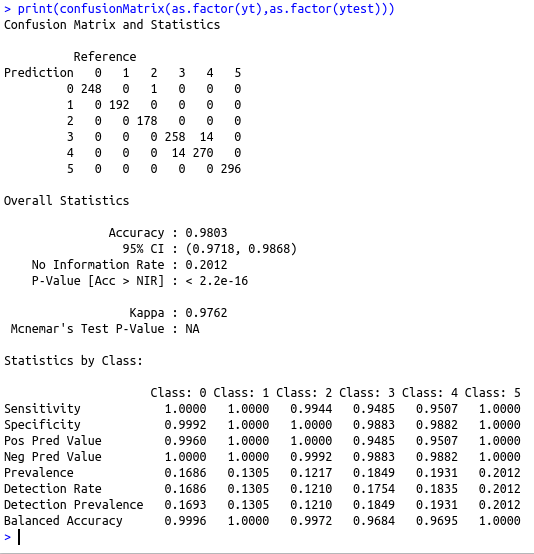
cat(“The confusion matrix is \n”)
print(confusionMatrix(as.factor(yt),as.factor(ytest)))