To choose the best fit Classification Model for the given data set using R.
#Import Data
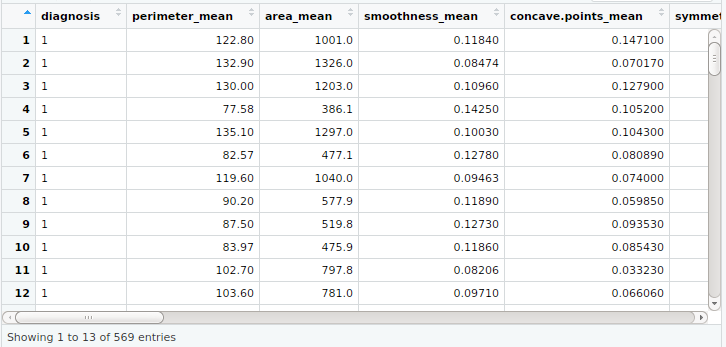
my_input<-read.csv(“data.csv”)
View(my_input)
str(my_input)
#Data Preparation
#Number of Missing Values
sum(is.na(my_input))
#List of rows having Missing Values
my_input[!complete.cases(my_input),]
#Dropping Missing Value variable and id
my_input<-my_input[,c(-1,-33)]
#Checking for Missing Values
sapply(my_input, function(x)sum(is.na(x)))
#Descriptive Statictics
summary(my_input)
#Univariate Plotting
#Pie Chart
tab<-table(my_input$diagnosis)
colors<-terrain.colors(2)
p_tab<-round(prop.table(tab)*100, digits = 2)
prop_tab<-as.data.frame(p_tab)
p_tab
lab<-sprintf(“%s – %3.1f%s”,prop_tab[,1],p_tab,”%”)
pie(p_tab,col =colors,labels = lab, clockwise = T, border = “gainsboro”,
radius = 1,cex=1.5,main = “Pie Chart of Frequency of Cancer Diagnosis”)
legend(“topright”,legend = prop_tab[,1],fill=colors,cex=1)
#Box Plot
colnames(my_input)
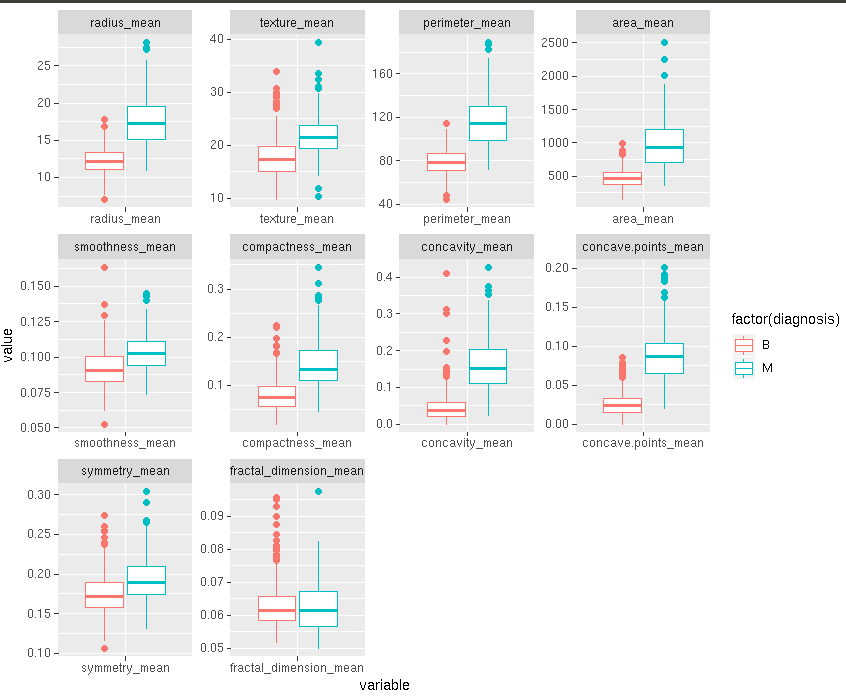
my_mean<-my_input[,c(1:11)];
my_se<-my_input[,c(1, 12:21)];
my_worst<-my_input[,c(1, 22:31)]
library(“ggplot2”)
library(“reshape2″)
ggplot(melt(my_mean,id.vars=”diagnosis”)) + geom_boxplot(aes(variable,value,color =factor(diagnosis))) +
facet_wrap(~variable, scales =’free’)
ggplot(melt(my_se,id.vars=”diagnosis”)) + geom_boxplot(aes(variable,value,color =factor(diagnosis))) +
facet_wrap(~variable, scales =’free’)
ggplot(melt(my_worst,id.vars=”diagnosis”)) + geom_boxplot(aes(variable,value,color =factor(diagnosis))) +
facet_wrap(~variable, scales =’free’)
#Bivariate Analysis
#install.packages(“corrplot”)
library(“corrplot”)
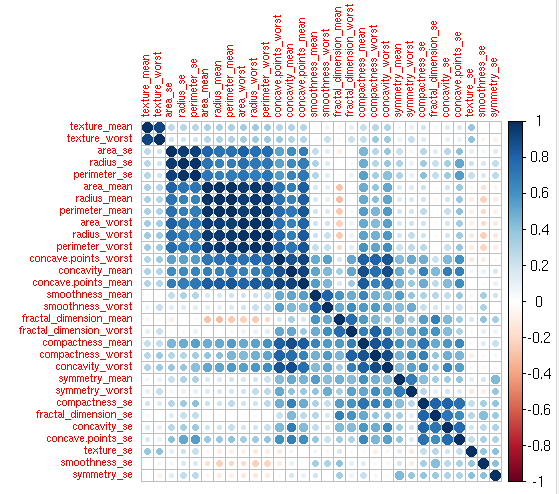
cor_val<-cor(my_input[,2:31])
corrplot(cor_val,order = “hclust”,tl.cex=0.7,main=”Correlation in Independent Variables”)
#Variables with High Correlation
#install.packages(“caret”)
library(“caret”)
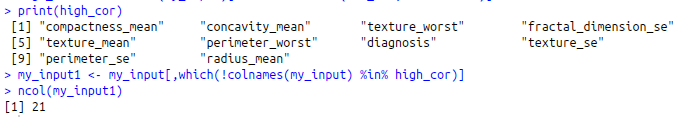
high_cor<-colnames(my_input)[findCorrelation(cor_val, cutoff=0.9)]
print(high_cor)
#Removing the highly correlated independent variables
my_input1<-my_input[,which(!colnames(my_input) %in% high_cor)]
ncol(my_input1)
#Splitting into train and test
levels(my_input$diagnosis)
input<-cbind(diagnosis=my_input$diagnosis,my_input1)
View(input)
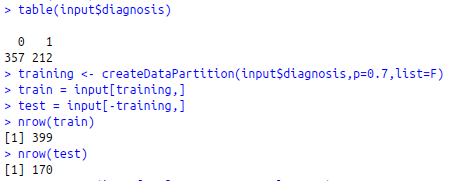
table(input$diagnosis)
training<-createDataPartition(input$diagnosis,p=0.7,list=F)
train = input[training,]
test = input[-training,]
nrow(train)
nrow(test)
#ML Algorithms
#Setting levels for both train and test data set
levels(train$diagnosis)<-make.names(levels(factor(train$diagnosis)))
levels(test$diagnosis)<-make.names(levels(factor(test$diagnosis)))
fit<-trainControl(method=”cv”,
number = 5,
preProcOptions = list(thresh = 0.99),
classProbs = TRUE,
summaryFunction = twoClassSummary)
#Logistic Regression
log_data<-glm(diagnosis~.,data=train,family = “binomial”)
summary(log_data)
log_data_train<-train(diagnosis~., data=train, method=”glm”,family=binomial(),
trControl=fit )
#ROC curve
#install.packages(“ROCR”)
library(“ROCR”)
pred<-prediction(predict(log_data,test),test$diagnosis)
per<-performance(pred,”tpr”,”fpr”)
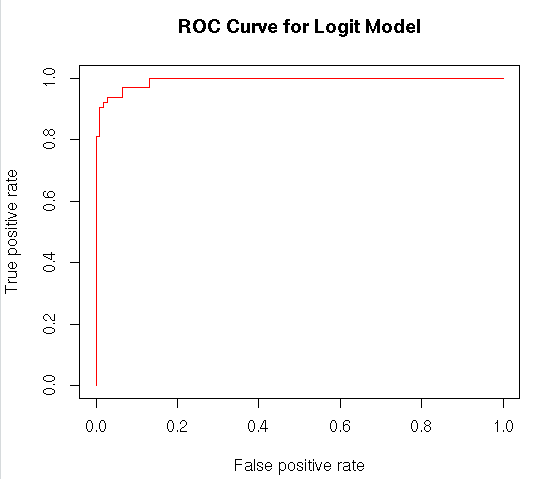
plot(per,col=”red”,main=”ROC Curve for Logit Model”)
#Prediction
predict_log<-round(predict(log_data,test,type = “response”),digits = 0)
predict_log<-as.factor(predict_log)
library(“caret”)
levels(predict_log)
levels(test$diagnosis)<-0:1
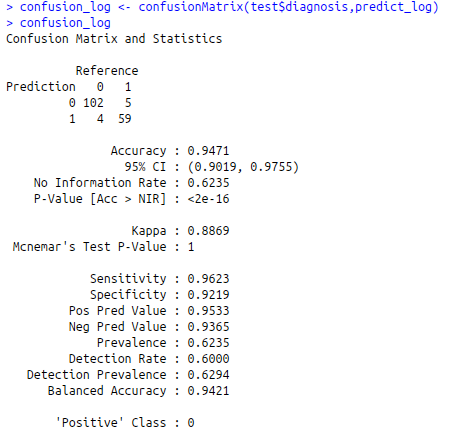
confusion_log<-confusionMatrix(test$diagnosis,predict_log)
confusion_log
#Random Forest
#install.packages(“randomForest”)
library(“randomForest”)
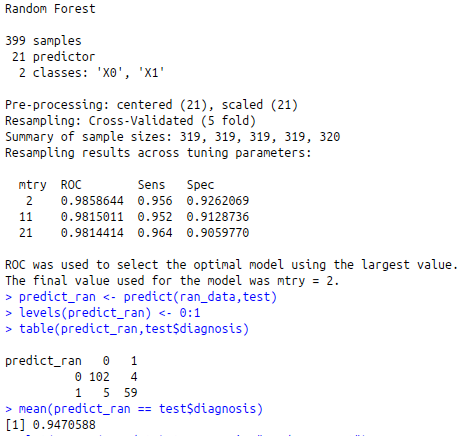
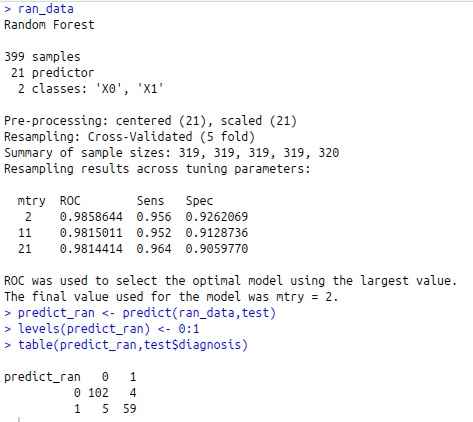
ran_data trControl=fit )
ran_data
predict_ran<-predict(ran_data,test)
levels(predict_ran)<-0:1
table(predict_ran,test$diagnosis)
mean(predict_ran == test$diagnosis)
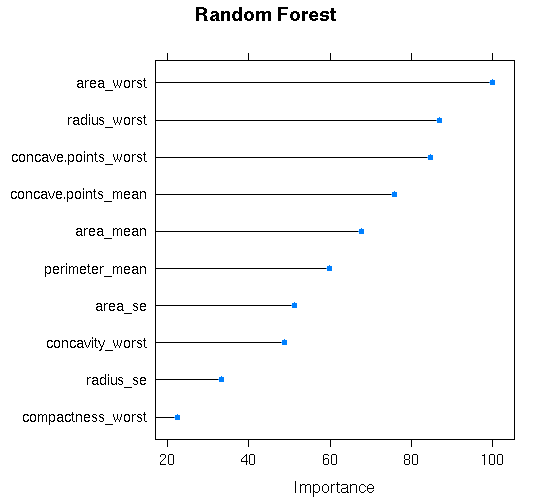
#Feature Importance Plot
plot(varImp(ran_data),top=10,main=”Random Forest”)
#Confusion Matrix
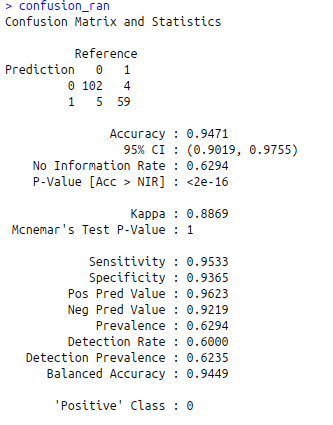
confusion_ran<-confusionMatrix(predict_ran,test$diagnosis)
confusion_ran
#Random Forest with PCA
ran_pca<-train(diagnosis~.,
data = train,
metric=”ROC”,
preProcess = c(‘center’, ‘scale’, ‘pca’),
trControl=fit)
#Prediction
predict_ran_pca<-predict(ran_pca, test)
levels(predict_ran_pca)<-0:1
#Confusion Matrix
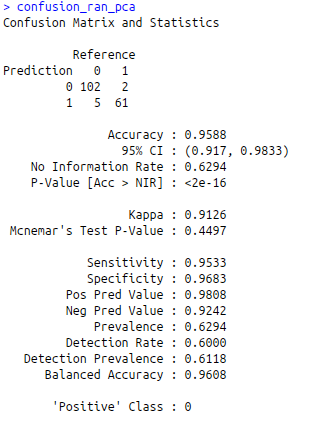
confusion_ran_pca<-confusionMatrix(predict_ran_pca, test$diagnosis)
confusion_ran_pca
#KNN
# Setting up train controls
library(“caret”)
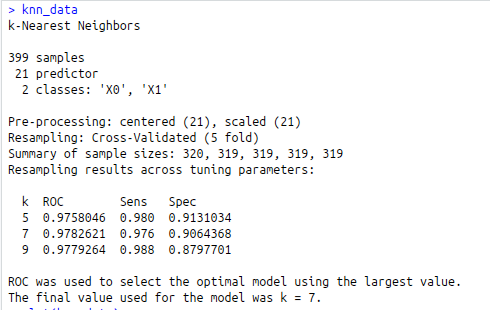
knn_data<-train(diagnosis~. , data = train, method = “knn”,
preProcess = c(“center”,”scale”),
trControl = fit,
metric = “ROC”)
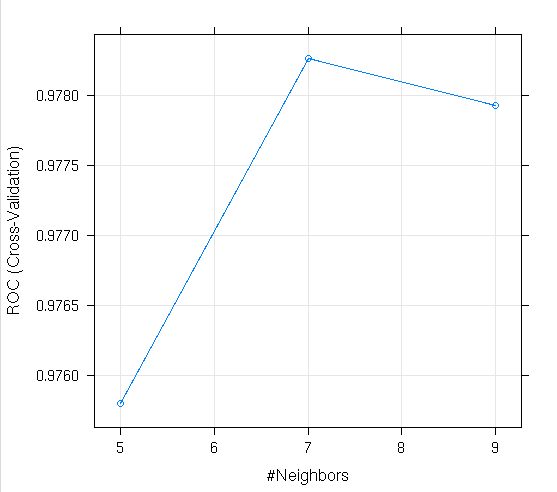
knn_data
plot(knn_data)
#Prediction
predict_knn<-predict(knn_data,test)
levels(predict_knn)<-0:1
#Confusion Matrix
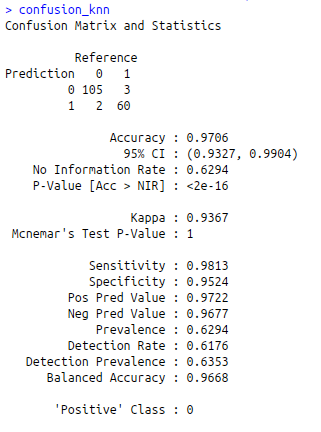
confusion_knn<-confusionMatrix(predict_knn,test$diagnosis)
confusion_knn
#SVM
library(“e1071″)
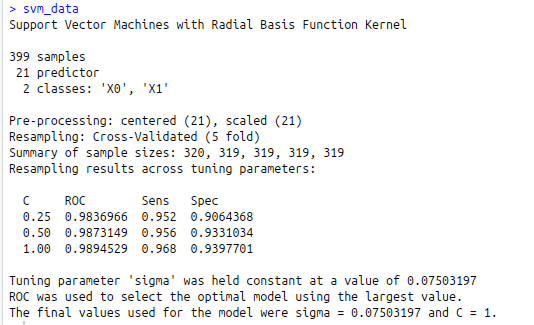
svm_data<-train(diagnosis~., data=train, method=”svmRadial”, metric=”ROC”, preProcess=c(‘center’,’scale’),
trace=F,trControl=fit )
svm_data
#Prediction
predict_svm<-predict(svm_data,test)
levels(predict_svm)
#Confusion Matrix
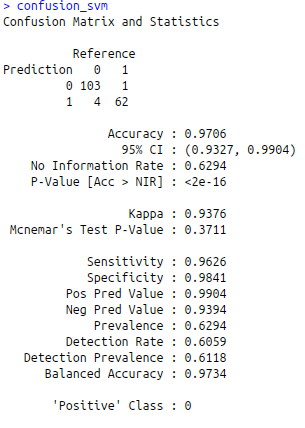
confusion_svm<-confusionMatrix(predict_svm,test$diagnosis)
confusion_svm
#Models Evaluation
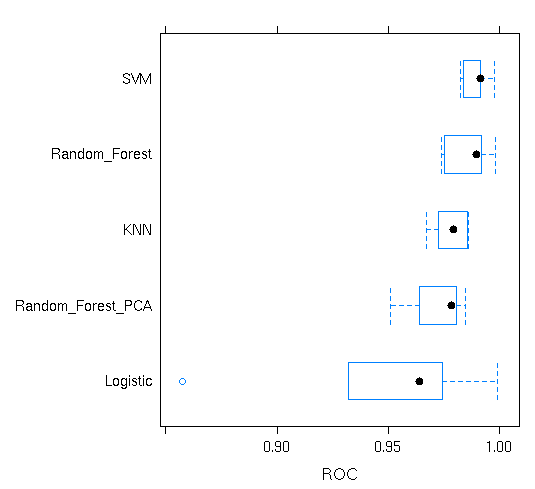
model_list<-list(Logistic=log_data_train,Random_Forest=ran_data ,Random_Forest_PCA=ran_pca ,KNN=knn_data,SVM=svm_data)
re
bwplot(re,metric = “ROC”)
confusion<-list(Logistic=confusion_log,Random_Forest=confusion_ran,Random_Forest_PCA=confusion_ran_pca,KNN=confusion_knn,SVM=confusion_svm)
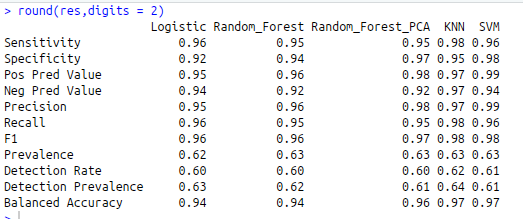
res<-sapply(confusion, function(x) x$byClass)
round(res,digits = 2)